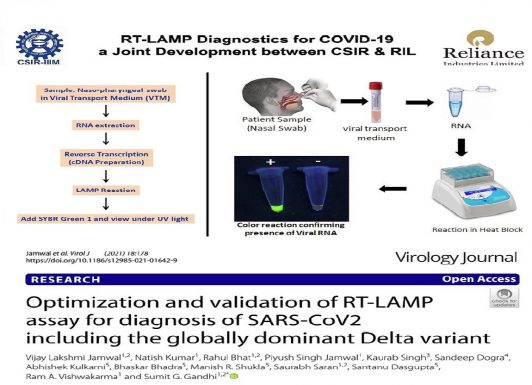
Abstract Background: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 pandemic, has infected more than 179 million people worldwide. Testing of infected individuals is crucial for identification and isolation, thereby preventing further spread of the disease. Presently, Taqman™ Reverse Transcription Real Time PCR is considered gold standard, and is the most common technique used for molecular testing of COVID-19, though it requires sophisticated equipments, expertise and is also relatively expensive.
Objective: Development and optimization of an alternate molecular testing method for the diagnosis of COVID-19, through a two step Reverse Transcription Loop-mediated isothermal AMPlification (RT-LAMP).
Results: Primers for LAMP were carefully designed for discrimination from other closely related human pathogenic coronaviruses. Care was also taken that primer binding sites are present in conserved regions of SARS-CoV2. Our analysis shows that the primer binding sites are well conserved in all the variants of concern (VOC) and variants of interest (VOI), notified by World Health Organization (WHO). These lineages include B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.427/B.1.429, P.2, B.1.525, P.3, B.1.526 and B.1.617.1. Various DNA polymerases with strand displacement activity were evaluated and conditions were optimized for LAMP amplification and visualization. Different LAMP primer sets were also evaluated using synthetic templates as well as patient samples.
 Screen Reader Access
Screen Reader Access